Introduction
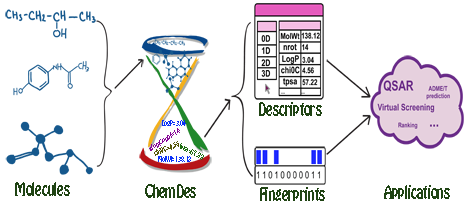
Molecular descriptors are experimentally-measured or theoretically-derived properties of a molecule. More specifically, they are quantitative representations of physical, chemical or topological characteristics of molecules that summarize our knowledge and understanding of molecular structure and activity from different aspects. Molecular fingerprints are property profiles of a molecule, usually in forms of bit or count vectors with the vector elements indicating the existence or the frequencies of certain properties, respectively. Both molecular descriptors and fingerprints play a fundamental role in QSAR/SAR analysis, virtual molecule screening, similarity-based compound search, target molecule ranking, drug ADME/T prediction and the other drug discovery processes.
ChemDes is a free web-based platform for the calculation of molecular descriptors and fingerprints, which provides more than 3,679 molecular descriptors that are divided into 61 logical blocks.In addition, it provides 59 types of molecular fingerprint systems for drug molecules, including topological fingerprints, electro-topological state (E-state) fingerprints, MACCS keys, FP4 keys, atom pairs fingerprints, topological torsion fingerprints and Morgan/circular fingerprints, et al.
Users that study on the characterization of various complex biological molecules and interaction samples, such as chemicals, proteins, DNA/RNA, and their interactions, can use another platform: BioTriangle
Please cite
Jie Dong, Dong-Sheng Cao, Hong-Yu Miao, Shao Liu, Bai-Chuan Deng, Yong-Huan Yun, Ning-Ning Wang, Ai-Ping Lu, Wen-Bin Zeng, Alex Chen. ChemDes: an integrated web-based platform for molecular descriptor and fingerprint computation. Journal of Cheminformatics 2015, 7:60
Molecular descriptors:
ChemDes allows users to compute 3679 molecular descriptors from several open source packages (for details see Molecular descriptors library).
Molecular fingerprints:
ChemDes allows users to compute 59 types of molecular fingerprints (for details see Molecular fingerprints library).
Tools
ChemCONV: Molecular format CONVersion of Chemicals.
ChemMOP: Molecular optimization of Chemicals based on MOPAC.
ChemFPS: Molecular Similarity of Chemicals based on FingerPrints.
- Version 1.0(2013-10-20). The ChemDes Version 1.0 was released
- Version 1.1(2013-11-20). Added module Molecular Fingerprints.Added the following fingerprints(topological fingerprints, electro-topological state (E-state) fingerprints, MACCS keys, FP4 keys, atom pairs fingerprints, topological torsion fingerprints and Morgan/circular fingerprints.)
- Version 1.1.1(2013-11-27).Fixed bugs.optimized the generating process of the csv file.
- Version 1.2(2013-12-05). Added tool:ChemCONV.Users can use this to convert the format of molecule and prepare the inputformats
- Version 1.3(2014-02-07). Added tool:ChemMOP.It is developed based on PyBel and MOPAC2012 to help studying of molecular structures,
- Version 1.4(2014-03-16). Added tool:ChemFPS.It is a free online-tool developed for Calculating Molecular Similarity based on molecular fingerprints by using measures such as 'Tanimoto'.
- Version 1.5(2014-07-11). Added new descriptors from CDK.
- Version 1.6(2014-08-18). Added new descriptors from RDKit.
- Version 1.7(2014-10-06). Added new descriptors from Pybel.
- Version 1.8(2014-11-11). Added new descriptors from BlueDesc.
- Version 1.9(2014-11-17). Added new descriptors from PaDEL.
- Version 2.0(2014-11-23). Update the tool ChemFPS. Added the batch computing function.
- Version 3.0(2014-12-01). Update the module Molecular Fingerprints . Added the following fingerprints('FP2 fingerprints', 'FP3 fingerprints', 'RDKit fingerprints', 'Pattern fingerprints', 'Layered fingerprints', 'Pubchem fingerprints', 'CDK fingerprints', 'CDK extended fingerprints' 'Klekota-Roth fingerprints', 'Klekota-Roth fingerprint count', 'CDK graph only fingerprints', 'Substructure fingerprints', 'Substructure fingerprint count', '2D atom pairs', '2D atom pairs count', 'Hybridization fingerprints'.).
- Version 3.0(2016-11-09). Fixed bugs and updated MOPAC to MOPAC2016.
- Version 3.0(2016-12-26). Fixed JSME Editor link.
- Version 3.0(2016-11-02). The ChemMOP service is temporary offline for the MOPAC update.
- Version 3.0(2016-12-15). The ChemMOP service is resumed.
- Version 3.1(2020-02-20). Fixed bug in molecular 'draw' editor.
- Version 3.2(2020-07-14). Fixed the display error in ChemCONV.
- The source code of the whole project is avaliable here.
- ChemDes by CBDD Group, CSU, China. is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Based on a work at http://www.scbdd.com/chemdes.
Permissions beyond the scope of this license may be available at http://www.scbdd.com/chemdes. If you have any questions, please feel free to contact us.